Resources
Next generation nanoLC-MS/MS-based proteomics on high-resolution, high-speed mass spectrometry platforms allows for unbiased profiling of protein (variant) expression, and may substantially expand our ability to understand the association between cancer-related genomic variation and cancer phenotypes.
Infrastructure
The current infrastructure consists of two nano-liquid
chromatography systems on-line coupled to tandem mass spectrometers:
EVosep-TimsTOF-HT (Bruker) and a QExactive HF platform.
In addition, two first generation QExactives are
available and a QTrap 5500.

Tandem mass spectrometers at the OPL (CCA 1-47)
IT Infrastructure
After shot-gun data acquisition by nanoLC-MS/MS, raw data are processed by the software tool MaxQuant for peptide/protein identification and quantification. For DIA-MS data we use DIA-NN/Spectronaut in conjunction with dedicated libraries.
Data exports to Excel are used for further dedicated statistical analyses, which is also facilitated by the OPL.
The computer infrastructure to cope with the large data flows is regularly upgraded. Currently, computing servers with a total of about 100 cores are connected to the tandem MS data acquisition PCs for fast data processing. Storage servers provide hundreds of TB of local, temporary storage. These servers are connected to the VUmc store4ever system for long term archive, as well as VUmc GreenQloud and the national computing grid for additional computing power.
 Forms & Protocols
Forms & Protocols
- OPL Sample Submission Sheet – nanoLC-MS/MS (form to be filled out electronically; a financial cost center/payment reference on row 2 is obligatory)
- OPL Sample and Data logging companion
- Preparation of Cell Line Lysates
- Tissue Lysate Preparation
- Bio-Rad Gel electrophoresis
- Coomassie Staining&Gel Storage
- Slice-By-Slice IGD protocol, with a supplementary figure.
- Whole-gel IGD
- Example gel image

Video instructions
Watch Instruction video FFPE proteomics (English) on Vimeo: https://vimeo.com/262529622
Watch cutting bones 2018 on Vimeo: https://vimeo.com/257870862
Watch video biobank (short) on Vimeo: https://vimeo.com/271285775
Watch video biobank (long) on Vimeo: https://vimeo.com/271285797
Web tool
Please cite the following papers when using the tool:
Beekhof R, van Alphen C, Henneman AA, Knol JC, Pham TV, Rolfs F, Labots M, Henneberry E, Le Large TY, de Haas RR, Piersma SR, Vurchio V, Bertotti A, Trusolino L, Verheul HM, Jimenez CR. INKA, an integrative data analysis pipeline for phosphoproteomic inference of active kinases. Mol Syst Biol. 2019 Apr 12;15(4):e8250. doi: 10.15252/msb.20188250. PubMed PMID: 30979792; PubMed Central PMCID: PMC6461034.
Software
Please cite the following papers when using our tests:
Pham TV, Jimenez CR. Simulated linear test applied to quantitative proteomics. Bioinformatics. 2016 Sep 1;32(17):i702-i709.
Pham TV, Jimenez CR. An accurate paired sample test for count data. Bioinformatics. 2012 Sep 15;28(18):i596-i602.
Pham TV, Piersma SR, Warmoes M, Jimenez CR. On the beta-binomial model for analysis of spectral count data in label-free tandem mass spectrometry-based proteomics. Bioinformatics. 2010 Feb 1;26(3):363-9.
Education
The OPL organizes every year an optional VUmc master course entitled: ‘Biomedical Proteomics’ of 2 weeks that runs end of January-early February.
It encompasses one week of theory (lectures) and one week of practical work as well as hands-on training in data analysis.
It is intended for VUmc master students and our collaborators who wish to obtain more in-depth knowledge of proteomics. Collaborators often bring their own sample for a pilot analysis.
Click here for an impression of the 2018 course https://quik.gopro.com/v/nCPCXMVUV4/
Pictures of the 2020 course


Tutorial Figures
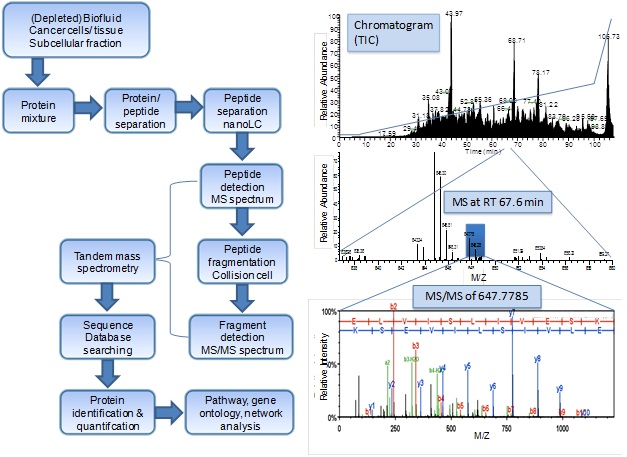
Protein identification by mass spectrometry-based proteomics
Biomarker discovery and validation pipeline
Process flow for the development of novel biomarker candidates. Each of the five biomarker development phases has its specific aims and requirements. Experiment size refers to the numbers of proteins expected to be evaluated as candidate biomarkers in each phase of development in relation to the sample requirements. Adapted from Suriniva et al., J.Proteome.Res. 10(1), 5-16 (7-1-2011).
OPL Meetings
Every Friday 14.00-~15.30/16.00.
All lab members present updates on their research. Collaborators are welcome to join in and present their work as well.
Journal club
Bi-weekly on Fridays 13.00-14.00 and sometimes on another day.
Medical Oncology Dept. seminar:
Every Friday 16.00-17.00.
CCA NeXt and CCA seminars
Ad hoc
